One
of the more popular arguments used for humans supposedly evolving from
apes is known as the chromosome fusion. The impetus for this concept is
the evolutionary problem that apes have an extra pair of
chromosomes—humans have 46 while apes have 48. If humans evolved from an
ape-like creature only three to six million years ago, a mere blip in
the grand scheme of the evolutionary story, why do humans and apes have
this discrepancy?
One
of the more popular arguments used for humans supposedly evolving from
apes is known as the chromosome fusion. The impetus for this concept is
the evolutionary problem that apes have an extra pair of
chromosomes—humans have 46 while apes have 48. If humans evolved from an
ape-like creature only three to six million years ago, a mere blip in
the grand scheme of the evolutionary story, why do humans and apes have
this discrepancy?
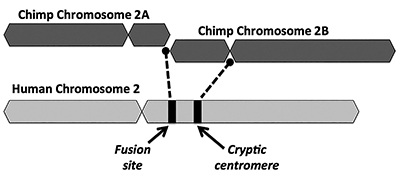
The evolutionary solution proposes that an end-to-end fusion of two small ape-like chromosomes (named 2A and 2B) produced human chromosome 2 (Figure 1). The concept of a fusion first came about in 1982 when scientists examined the similarities of human and ape chromosomes under a microscope. While the technique was somewhat crude, it was enough to get the idea going.1
The So-Called Fusion Site
The first actual DNA signature of a possible fusion event was discovered in 1991 on human chromosome number 2.2 Researchers found a small, muddled cluster of telomere-like end sequences that vaguely resembled a possible fusion. Telomeres are a six-base sequence of the DNA letters TTAGGG repeated over and over again at the ends of chromosomes.
However, the fusion signature was somewhat of an enigma based on the real fusions that occasionally occur in nature. All documented fusions in living animals involve a specific type of sequence called satellite DNA (satDNA) located in chromosomes and found in breakages and fusions.3-5 The fusion signature on human chromosome 2 was missing this telltale satDNA.6
Another problem is the small size of the fusion site, which is only 798 DNA letters long. Telomere sequences at the ends of chromosomes are 5,000 to 15,000 bases long. If two chromosomes had fused, you should see a fused telomere signature of 10,000 to 30,000 bases long—not 798.
Not only is the small size a problem for the fusion story, the signature doesn’t really represent a clear-cut fusion of telomeres. Figure 2 shows the DNA letters of the 798-base fusion site with the six-base (DNA letter) intact telomere sequences emphasized in bold print. When the fusion sequence is compared to that of a pristine fusion signature of the same size, it is only 70% identical overall.

Secular researchers have pointed out this discrepancy and have labeled the fusion site as significantly “degenerate.”7 Given the standard theoretical model of human evolution, it should be about 98 to 99% identical, not 70%. The researchers describing this discovery commented, “Head-to-head arrays of repeats at the fusion site have degenerated significantly (14%) from the near perfect arrays of (TTAGGG)n found at telomeres” and asked the pertinent question “If the fusion occurred within the telomeric repeat arrays less than ~6 Mya, why are the arrays at the fusion site so degenerate?”7 It should be noted that the 14% degeneration cited by the authors refers to the corruption of just the six-base sequences themselves, not the whole 798 bases.
The Fusion Site Inside a Gene?
The most remarkable anti-evolutionary finding about the fusion site turned out to be its location and what it actually does. This discovery came about while I was reading the research paper that reported a detailed analysis of 614,000 bases of DNA sequence surrounding the alleged fusion site. I noticed in one of the figures that the fusion site was located inside a gene, and quite remarkably this oddity wasn’t even acknowledged in the text of the paper.8
A finding like this is highly noteworthy. Perhaps this piece of information would’ve been the nail in the evolutionary coffin, so to speak, which is why the researchers declined to discuss it. This major anomaly inspired me to give the fusion site a much closer examination. This paper was published in 2002, and I took notice of it in 2013. A huge amount of data on the structure and function of the human genome had been published in the meantime, and there was likely much more to the story that needed to be uncovered.
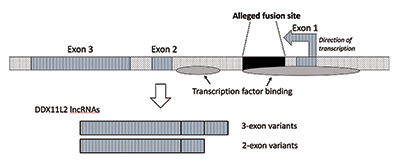
When I performed further research, I verified that the fusion site was positioned inside an RNA helicase gene now called DDX11L2. Most genes in plants and animals have their coding segments in pieces called exons so they can be alternatively spliced. Based on the addition or exclusion of exons, genes can produce a variety of products. The intervening regions between exons are called introns, which often contain a variety of signals and switches that control gene function. The alleged fusion site is positioned inside the first intron of the DDX11L2 gene (Figure 3).9
The DNA molecule is double-stranded, with a plus strand and a minus strand. It was engineered this way to maximize information density while also increasing efficiency and function. As a result, there are genes running in different directions on the opposing strands. As it turns out, the DDX11L2 gene is encoded on the minus strand. Because genes in humans are like Swiss army knives and can produce a variety of RNAs, in the case of the DDX11L2 gene it produces short variants consisting of two exons and long variants with three (Figure 3).9
The Fusion Site Is a Gene Promoter
What might this DDX11L2 gene be doing? My research showed it’s expressed in at least 255 different cell or tissue types.9 It’s also co-expressed (turned on at the same time) with a variety of other genes and is connected to processes associated with cell signaling in the extracellular matrix and blood cell production. The location of the so-called fusion sequence inside a functional gene associated with the genetics of a variety of cellular processes strongly refutes the idea that it’s the accidental byproduct of a head-to-head telomeric fusion. Genes are not formed by catastrophic chromosomal fusions!
Even more amazing is that the fusion site is itself functional and serves an important engineered purpose. The site actually acts as a switch for controlling gene activity. In this respect, a wealth of biochemical data showed that 12 different proteins called transcription factors regulate this segment of the gene. One of these is none other than RNA polymerase II, the main enzyme that copies RNA molecules from DNA in a process called transcription. Further supporting this discovery is the fact that the actual process of transcription initiates inside the region of the so-called fusion site.
Technically, we would call the activity in the alleged fusion site a promoter region. Promoters are the main switches at the beginning of genes that turn them on and are also where the RNA polymerase starts to create an RNA. Many genes have alternative promoters like the DDX11L2 gene.
There are actually two areas of transcription factor binding in the DDX11L2 gene. The first is in the promoter directly in front of the first exon, and the second is in the first intron corresponding to the fusion site sequence. Not only is the DDX11L2 gene itself complexly controlled, with the alleged fusion sequence playing a key role, but even the RNA transcripts produced are very intricate. The RNAs themselves contain a wide variety of binding and control sites for a class of small regulatory molecules called microRNAs.9
Functional Internal Telomere Sequences Are All Over the Genome
The presence of internally located telomere sequence is found all over the human genome. These seemingly out-of-place telomere repeats have been dubbed interstitial telomeres. The presence of these sequences presents another challenge for the fusion site idea. It’s a fact that very few of the telomere repeats in the fusion site occur in tandem. As noted in Figure 2, the sequence of the 798-base fusion site contains only a few instances where two repeats are actually in tandem and none that have three repeats or more. However, there are many other interstitial telomere sites all over the human genome where the repeats occur in perfect tandem three to ten times or more.10-11
Even besides their role at the ends of chromosomes, it appears interstitial telomeric repeats may serve an important function in the genome related to gene expression. In a recent research project, I identified telomere repeats all over the human genome and then intersected their genomic locations with a diversity of data sets containing functional biochemical information for gene activity.12 Literally thousands of internal telomeric repeats across the genome were directly associated with the hallmarks of gene expression. The same type of transcription factor binding and gene activity occurring at the alleged fusion site was also occurring genome-wide at numerous other interstitial telomeric repeats. Clearly, these DNA features are not accidents of evolution but purposefully and intelligently designed functional code.
Bogus Cryptic Centromere Inside a Gene
Another key problem with the fusion model is the lack of viable evidence for a signature of an extra centromere region. Centromeres are sections of chromosomes, often in central locations, that play key roles during cell division. As depicted in Figure 1, the newly formed chimeric chromosome would’ve had two centromere sites immediately following the alleged head-to-head fusion of the two chromosomes. In such a case, one of the centromeres would be functional while the other would be disabled. The presence of two active centromeres is bad news for chromosomes and would lead to dysfunction and cell destruction.
Interestingly, the evidence for a cryptic (disabled) centromere on human chromosome 2 is even weaker than that for a telomere-rich fusion site. Evolutionists explain the lack of a clearly distinguishable nonfunctional secondary centromere by arguing that a second centromere would’ve been rapidly selected against. After that, the disabled centromere would’ve deteriorated over time since there were no functional restraints placed on it anymore by its doing something useful in the genome.
However, the evidence for a second remnant centromere at any stage of sequence degeneration is problematic for the evolutionary paradigm. Functional centromere sequences are composed of a repetitive type of DNA called alphoid sequences, with each alphoid repeat being about 171 bases long. Some types of alphoid repeats are found all over the genome, while others are specific to centromeres. The structure of the sequences found at the cryptic centromere site on human chromosome 2 doesn’t match those associated with functional human centromeres.13 Even worse for the evolutionary model is that they have no highly similar counterparts in the chimp genome—they are human-specific.13
The alleged fossil centromere is also exceptionally tiny compared to a real one. The size of a normal human centromere ranges in length between 250,000 and 5,000,000 bases.14 The alleged cryptic centromere is only 41,608 bases long, but it’s also important to note that there are three different regions of it that aren’t even alphoid repeats.15 Two of these are called retroelements, with one being a LPA3/LINE repeat 5,957 bases long and the other an SVA-E element with 2,571 bases. When we subtract the insertions of these non-alphoid sequences, it gives a length of only 33,080 bases, which is a fraction of the length of a real centromere.
The most serious evolutionary problem with the idea of a fossil centromere, though, is that like the alleged fusion site, it’s positioned inside a gene. The alleged cryptic centromere is located inside the ANKRD30BL gene, and its sequence spans both intron and exon regions of the gene.12,15
In fact, the part of the alleged fossil centromere sequence that lands inside an exon actually codes for amino acids in the resulting gene’s protein. The gene produces a protein that’s believed to be involved in the interaction of the structural network of proteins inside the cell called the cytoskeleton in connection with receptor proteins embedded in the cell membrane.16 The fact that the so-called fossil or cryptic centromere is a functional region inside an important protein-coding gene completely refutes the idea that it’s a defunct centromere.
Conclusion: No Fusion
Due to the muddled signatures and small sizes of the alleged fusion and fossil centromere sites, it’s highly questionable that their sequence was evolutionarily derived from an ancient chromosome fusion. Not only that, they represent functional sequence inside genes. The alleged fusion site is an important genetic switch called a promoter inside the DDX11L2 long noncoding RNA gene, and the so-called fossil centromere contains both coding and noncoding sequence inside a large ankyrin repeat protein-coding gene.
This is an undeniable double whammy against the whole mythical fusion idea, utterly destroying its validity. The overwhelming scientific conclusion is that the fusion never happened.
References
- Yunis, J. J. and O. Prakash. 1982. The origin of man: a chromosomal pictorial legacy. Science. 215 (4539): 1525-1530.
- Ijdo, J. W. et al. 1991. Origin of human chromosome 2: An ancestral telomere–telomere fusion. Proceedings of the National Academy of Sciences. 88 (20): 9051-9055.
- Chaves, R. et al. 2003. Molecular cytogenetic analysis and centromeric satellite organization of a novel 8;11 translocation in sheep: a possible intermediate in biarmed chromosome evolution. Mammalian Genome. 14 (10): 706-710.
- Tsipouri, V. et al. 2008. Comparative sequence analyses reveal sites of ancestral chromosomal fusions in the Indian muntjac genome. Genome Biology. 9 (10): R155.
- Adega, F., H. Guedes-Pinto, and R. Chaves. 2009. Satellite DNA in the Karyotype Evolution of Domestic Animals—Clinical Considerations. Cytogenetic and Genome Research. 126 (1-2): 12-20.
- Tomkins, J. P. and J. Bergman. 2011. Telomeres: implications for aging and evidence for intelligent design. Journal of Creation. 25 (1): 86-97.
- Fan, Y. et al. 2002. Genomic Structure and Evolution of the Ancestral Chromosome Fusion Site in 2q13–2q14.1 and Paralogous Regions on Other Human Chromosomes. Genome Research. 12 (11): 1651-1662.
- Fan, Y. et al. 2002. Gene Content and Function of the Ancestral Chromosome Fusion Site in Human Chromosome 2q13–2q14.1 and Paralogous Regions. Genome Research. 12 (11): 1663-1672.
- Tomkins, J. P. 2013. Alleged Human Chromosome 2 “Fusion Site” Encodes an Active DNA Binding Domain Inside a Complex and Highly Expressed Gene—Negating Fusion. Answers Research Journal. 6: 367-375.
- Azzalin, C. M., S. G. Nergadze, and E. Giulotto. 2001. Human intrachromosomal telomeric-like repeats: sequence organization and mechanisms of origin. Chromosoma. 110: 75-82.
- Ruiz-Herrera, A. et al. 2008. Telomeric repeats far from the ends: mechanisms of origin and role in evolution. Cytogenetic and Genome Research. 122 (3-4): 219-228.
- Tomkins, J. P. 2018. Combinatorial genomic data refute the human chromosome 2 evolutionary fusion and build a model of functional design for interstitial telomeric repeats. In Proceedings of the Eighth International Conference on Creationism. J. H. Whitmore, ed. Pittsburgh, PA: Creation Science Fellowship, 222-228.
- Tomkins, J. and J. Bergman. 2011. The chromosome 2 fusion model of human evolution—part 2: re-analysis of the genomic data. Journal of Creation. 25 (2): 111-117.
- Aldrup-Macdonald, M. E. and B. A. Sullivan. 2014. The Past, Present, and Future of Human Centromere Genomics. Genes (Basel). 5 (1): 33-50.
- Tomkins, J. P. 2017. Debunking the Debunkers: A Response to Criticism and Obfuscation Regarding Refutation of the Human Chromosome 2 Fusion. Answers Research Journal. 10: 45-54.
- Voronin, D. A. and E. V. Kiseleva. 2008. Functional Role of Proteins Containing Ankyrin Repeats. Cell and Tissue Biology. 49 (12): 989-999.
* Dr. Tomkins is Life Sciences Director at the Institute for Creation Research and earned his Ph.D. in genetics from Clemson University.